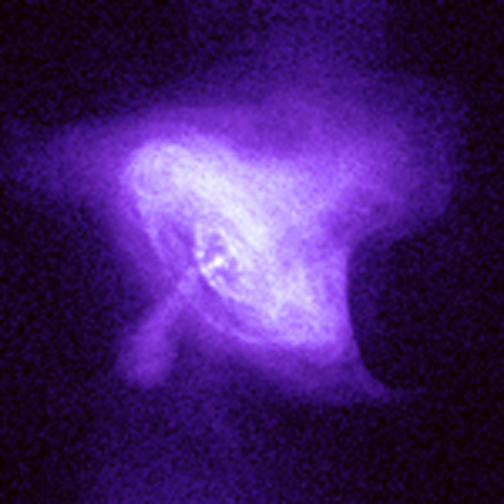
Last January Doc Muriel discussed something about laminar fluids emitting blue light and that turbulent fluids would emit at a lower wavelength. He showed a picture of the Crab Nebula and the decomposition to blue. The result showed a blue ring at the center of the Nebula. It surprised me that such phenomena as laminar fluids in space could be seen in a simple nebula image. I never saw it again.
I'd love to have a 7-band image of the nebula; all I have so far is a snapshot from Stellarium. I tried to get the blue channel. Of course it's an RGB image and the blue channel also contributes to form other colors.
Here is an attempt to further filter the blue channel by removing pixels that overlap higher level red and green (>0.7 with max 1.0). It's a crude attempt but I'll build on this and find an actual decomposition of the nebula for comparison.
Here are nice crab photos
x-rays from Crab Nebula
Here is the code.
A =imread('crab.png');
red = A(:,:,1);
green = A(:,:,2);
blue = A(:,:,3);
//red channel
R = A;
R(:,:,2) = 0;
R(:,:,3) = 0;
//green channel
G = A;
G(:,:,1) = 0;
G(:,:,3) = 0;
//blue channel
B = A;
B(:,:,1) = 0;
B(:,:,2) = 0;
scf(2); imshow(B)
scf(3); imshow(B(:,:,3));
//640-490nm
rc = R(:,:,1);
rc(find(rc>0.7)) = 0;
gc = G(:,:,2);
gc(find(gc>0.7)) = 0;
blue = B(:,:,3).*rc.*gc;
B(:,:,3) = blue;
scf(4);imshow(blue);
imwrite(blue, 'bluecrab.png');
imwrite(B(:,:,3), 'bluechannel.png');
imwrite(B, 'bluecrab3.png');